Please click the button below to go to our email login page
|
Details|Eight principles to design PCR primers1. Purposes for designing PCR primers PCR primers are designed for seeking a pair of appropriate nucleotide fragments to ensure that they can amplify template DNA sequences (target gene) with high quality. Thus, the quality of primers is crucial for the specificity of PCR and the success of experiments.
2. Eight principles for primer design
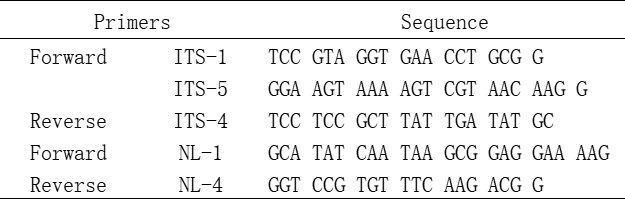
Principle 1: Ensure the specificity of primers To ensure the specificity of PCR amplification, the homology between the primers and the non-specific amplification sequences should not exceed 70%, or there should be at least 8 consecutive complementary base homology.
Principle 2: Select appropriate primer length The length of the oligonucleotide primers is 15 to 30 bp, normally 20~27 mer. The effective length of primers is Ln=2 (G+C)+(A+T), among which the value of Ln should not exceed 38. When the value of Ln>38, the most appropriate temperature for primer extension exceeds the most appropriate temperature (74°C) for TaqDNA polymerase, thereby failing to ensure the specificity of primers.
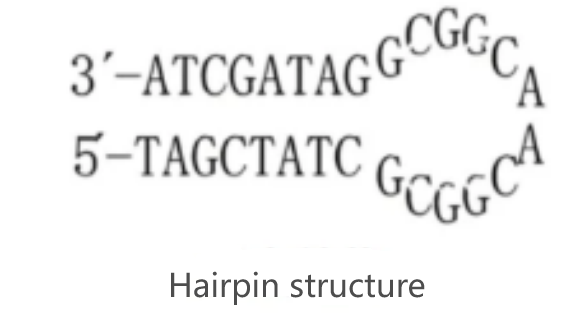
Principle 3: Ensure the product does not form secondary structures.
A primary reason for some primers ineffectiveness is the impacts from DNA secondary structures in primer repetitive regions. When selecting the amplification fragments,it’s necessary to avoid secondary structure regions.Assessing the stable secondary structure of mRNA with appropriate computer software is conducive to optimal template selection.
Principle 4: Ensure random distribution of bases
During primer design, the four bases should be distributed randomly to avoid accumulation of purines or pyrimidines. It is particularly important to note that the consecutive number of G or C at the 3’ end should not exceed 3, as this may cause errors of the primers in the G+C enriched sequence region.
Principle 5: Ensure an appropriate G+C content
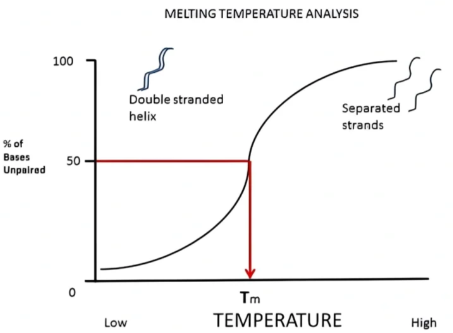
The optimal G+C content is usually 40–60 %. Tm is the melting temperature of oligonucleotides, at which 50 % of the oligonucleotide duplex dissociates under defined salt conditions. Temperature for efficient initiation typically is 5–10 °C higher than the Tm. Using the formula Tm = 4(G + C) + 2(A + T), Tm of effective primers is between 55 °C~80 °C. To achieve the optimal annealing conditions, the Tm values of the primers should be close to 72°C.
Principle 6: Avoid self- or inter-primer complementarity of ≥4 consecutive bases Besides, the two primers must not be complementary, especially at their 3’ ends, to prevent primer-dimer formation. Meanwhile, the homology or complementarity of consecutive bases between primers should be controlled, and should not exceed 4.
Principle 7: Modifications allowed at the 5’ end and prohibited at the 3’ end The 5’ end of primers defines PCR product length but has little influence on extension specificity, the modification of which therefore exerts no effect on extension specificity. The modification of the primers 5’ end includes the addition of restriction enzyme cutting site; labeled biotin, fluorophores, digoxigenin, Eu³⁺, etc.; introducing protein-binding DNA sequence; introducing mutation sites, inserting and deleting mutation sequences, and introducing a small segment of promoter sequence, etc.
The primer extension begins at the 3’ end and cannot be modified in any way. Moreover, there should be no possibility of forming secondary structures at the 3’ end. Except in special PCR (AS-PCR) reactions, mismatches should not occur at the 3’ end of the primer.
Principle 8: Ensure that the 3’ end of the primer evades the third position of the codon The 3’ end of a primer should not terminate at the third position of a codon, because this site is highly degenerate and can impact both specificity and efficiency of amplification. |