Please click the button below to go to our email login page
|
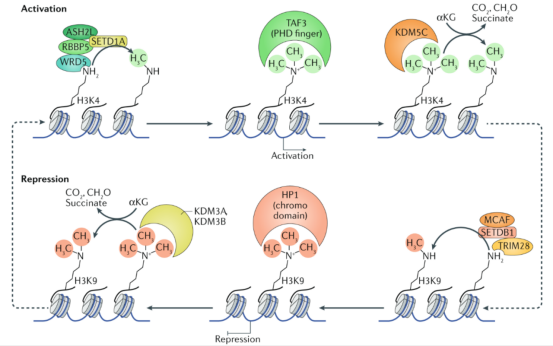
What are the types of histone modifications? What are their characteristics?It is known that chromosomes are the carrier of life genetic information, and DNA, LncRNA and histone modification can regulate gene transcription and expression. Of them, post-translational modification of histones mainly occurs on their N-terminal protein tails, and various modification types and sites constitute the diversity of histone function. 01. Naming of histones The main family of histone: H1/H5 (linker histones), H2A, H2B, H3, H4; Histone amino acid: K (lysine), R (arginine); Modification types: Me (methyl), P (phosphate), Ac (acetyl), Ub (ubiquitin). For example, H3K4me3 represents the trimethylation of histone H3 lysine 4. 02. Regulatory enzymes of histone modification Writers: modifying enzymes (adding modifying gene group); Erasers: demodifying enzymes (removing modifying gene group); Readers: recognition proteins (identifying modification information). In a research involving identification of histone modifying enzymes and their complexes, the first step is to evaluate histone modification level through Chromatin immunoprecipitation-sequencing (ChIP-seq), followed by RNA-seq analysis of the regulation of histone modification on gene expressions, so as to form a complete chain of evidence for the epigenetic modification mechanism. The regulation of histone modification on gene transcription can be classified into direct regulation and indirect regulation. (1) Direct regulation: Histone modification affects nucleosome-nucleosome interactions through inserting modification. (2) Indirect regulation: Histone acetyltransferases are recruited by transcription factors (TFs) or use their intrinsic bromodomains to bind to pre-existing acetylated lysine residues and forcefully acetylate histones at the promoter region. 03. Types of histone modification The main types of histone modification are histone phosphorylation, histone methylation, histone acetylation and histone ubiquitination. (1) Histone phosphorylation Histone phosphorylation can mark DNA damage sites, which is an important part of histone modification. Without repair markers, DNA will be damaged by accumulated detrimental impacts from sources such as solar ultraviolet radiation. γH2AX, as a binding site of MDC1, is a key protein to recruit DNA repair protein. (2) Histone methylation Histone methylation includes: Two amino acid modifications: lysine methylation, arginine methylation; Three states: monomethylation, dimethylation, trimethylation; Methylation regulatory enzymes: histone methyltransferase (HMT), histone demethylase (HDM). The support from methylation regulatory enzymes is necessary, since histone methylation is reversible.
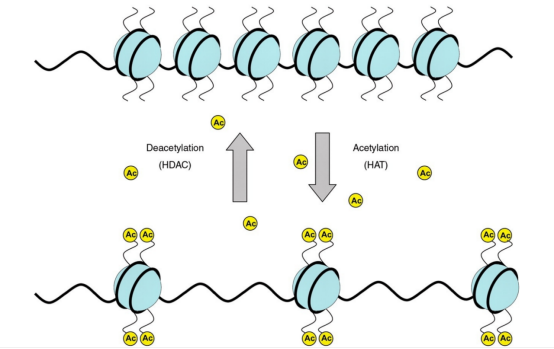
(3) Histone acetylation Histone acetylation is generally composed of: Acetylation reader: bromodomain, double PHD zinc finger structure, YEAST domain; Acetylation regulatory enzymes: histone lysine acetyltransferase, histone deacetylated transferase. Generally, the target protein is directly purified within cells, followed by mass spectrum identification to determine the acetylation site.
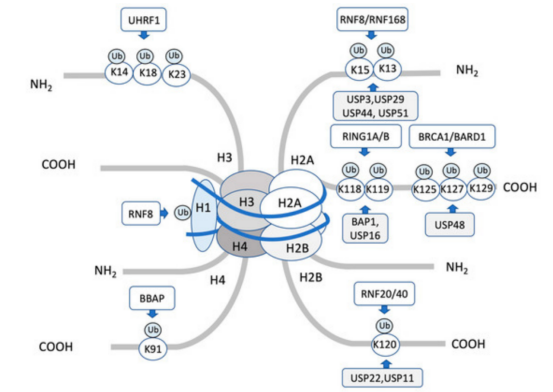
(4) Histone ubiquitination Histone ubiquitination is the process of binding the carboxyl-terminus of ubiquitin molecules to the lysine residues of proteins. Ubiquitination regulatory enzymes contain ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzymes (E2) and ubiquitin-protein ligases (E3). Histone ubiquitination can be influenced by several factors, such as: l Free ubiquitin available in cells, as well as the activity of histone ubiquitinases or deubiquitinating enzymes; l Histone ubiquitination occurs through an enzymatic cascade, involving E1, E2, E3 and other enzymes, while histone deubiquitination is triggered by peptidases.
|