Please click the button below to go to our email login page
|
How to conduct research on histone modification? Let’s unveil the research thought in paper with a high scoreThe research thought regarding histone modification can generally be divided into following types: (1) Comparison and identification of modification types, such as the finding of new modification, as well as competition, crosstalk and coordination of different modifications at the same site (2) Combination of existing modifications with mechanism and metastasis, such as research on the relation between histone modification with cancer development mechanism or inflammation mechanism (3) Biological function study (4) Effector protein study, such as the exploration of new modifications of effector proteins (5) Study on relevant modifying and demodifying enzymes, such as purification and identification of modifying enzymes Alexander van Oudenaarden, a group leader at Hubrecht Institute and a professor at the University Medical Center Utrecht, published a research paper, “scChIX-seq infers dynamic relationships between histone modifications in single cells” in Nature Biotechnology earlier this year.
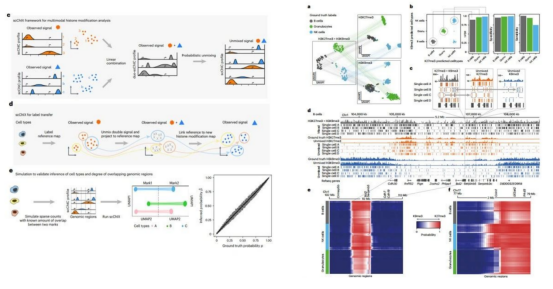
Let’s learn the research thought of histone modification in this paper. Research background: Gene expression in animals depends on epigenetic marks such as histone modifications to regulate the accessibility and function of the genome in different cell types. Chromatin immunoprecipitation followed by sequencing (ChIP–seq) based on enzyme family (chromatin immunocleavage, ChIC) can reduce the background signal in profiling the epigenome, and have enabled single-cell profiling of histone modifications. scChIX-seq links single-cell profiles of different histone modifications, revealing relationships between histone modifications in single cells and thus unlocking joint analysis of several histone modifications in single cells. Step 1 scChIX-seq can unbiasedly assign reads to each mark regardless of the amount of overlap between the two marks in the genome. Accordingly, the author unveils the relation between histone modifications in single cells through linking single-cell profiles of different histone modifications with scChIX-seq.
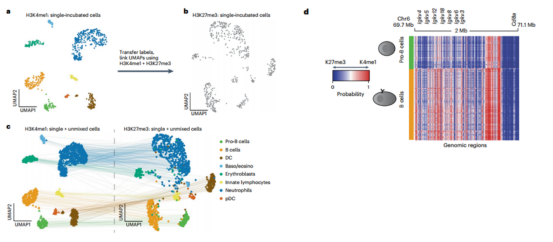
Step 2 To reveal the relation of H3K4me1 and H3K27me3 in bone marrow, the author sampled bone marrow from mice, and integrated active (H3K4me1) and repressive (H3K27me3) chromatins into cells to conduct joint analyses.
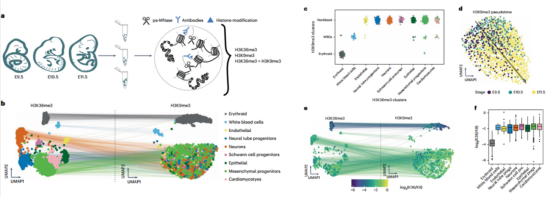
Step 3 Next, the author applied scChIX-seq to mouse organogenesis, and unraveled the difference between H3K36me3 and H3K9me3 in mouse organogenesis.
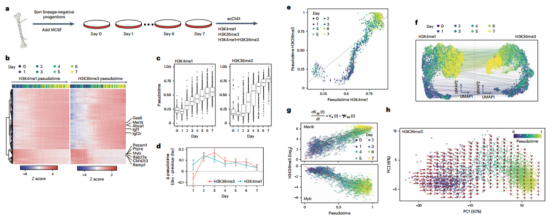
Step 4 Ultimately, the author studied the dynamic relationships between H3K4me1 and H3K36me3 over an in vitro differentiation timecourse.
This paper proved that scChIX-seq can deconvolve multiplexed histone modifications, expand the number of histone marks that can be analyzed in single cells, reveal biological insights by multimodal analysis, and unravel the data obscured by analyzing each modality separately. Here, we also want to recommend two other high-score literature, and if you are interested in histone modification, you can also search and read them. The first one is a paper pertaining to the relation of histone modification and diseases, which expounds the different effects of existing post-translational modifications of histones on various pathologies of human disease.
The second one is a paper regarding histone modification and its application, where ChIP, immunohistochemistry, immunofluorescence, qPCR, Transwell and Western blot assays were conducted to prove that FAK inhibitors can reverse breast cancer metastasis induced by upregulation of NEDD9 via pan-HDAC inhibitors, which may provide a potential combined treatment for breast cancer.
|