Please click the button below to go to our email login page
|
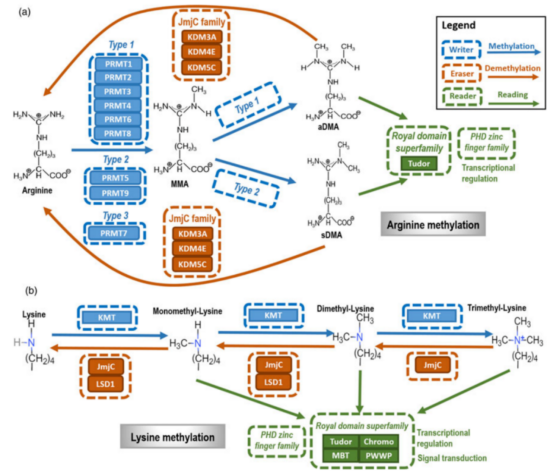
The starting point for research related to protein post-translational methylated modificationMethylation refers to the process in which proteins transfer methyl to and covalently bind to specific amino acid residues under the catalysis of methyltransferase. The following figure displays the main types and mechanisms of protein methylation. Protein methylation can occur in histones and non-histone proteins, and usually in arginine (Arg) and lysine (Lys) residues.
Figure A reveals that protein arginine methyltransferase (PRMT) triggers Arg methylation. In addition, members of the Jumonji C-terminal domain (JmjC) family, such as KDM3A, KDM4E and KDM5C, can serve as Arg demethylase to catalyze Arg demethylation. Tudor domain and PHD zinc finger domain, as Arg methylation-dependent binding proteins, can recognize Arg methylation, which plays various roles in different cells. Figure B indicates that Lys methylation is mediated by Lys-specific methyltransferase (KMT). Besides, JmjC family members and LSD1 are primary demethylases of Lys demethylation. The “Royal” superfamily domain and PHD zinc finger domain, constituted by Tudor, chromosome, MBT, and PWWP domains, are Lys methylation-dependent binding proteins.
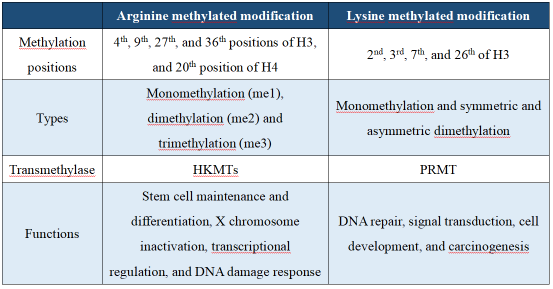
Conclusion of arginine and lysine methylation The research related to protein methylated modification can generally concentrate on whether the target protein has methylation or has multiple methylation positions, the interaction between different methylation types, and the mechanism of action of different methylases on the target protein. The following is a literature related to protein post-translational methylated modification with a high IF, titled PRMT3-mediated arginine methylation of IGF2BP1 promotes oxaliplatin resistance in liver cancer, which has been published in Nature Communications.
In this article, PRMT3 was first confirmed as the contributor of drug-resistant OXA in hepatocellular carcinoma (HCC) cells through functional screening using genome-wide CRISPR activation library, which also can promote OXA resistance in vivo and in vitro. Then, the molecular mechanism was further explored. It was proved that PRMT3 methylates IGF2BP1 at R452, and IGF2BP1 methylation is necessary for OXA resistance. Also, PRMT3 and IGF2BP1 m6A-dependently regulate HEG1 expression, the effect of which on OXA resistance has been experimentally verified to be dependent on HEG1, thus determining that the PRMT3-IGF2BP1-HEG1 axis is a regulatory factor for OXA resistance in HCC. The ultimate clinical correlation verification revealed that PRMT3 expression level is associated with poor clinical outcome of HCC patients and OXA-based HAIC treatment. This research obtained three conclusions: 1. PRMT3-IGF2BP1-HEG1 axis is a regulator of OXA resistance in HCC. 2. Targeting PRMT3 may be an effective method to improve the response of HCC cells to OXA treatment. 3. PRMT3 expression in biopsy specimens may predict patients’ response to OXA-based HAIC. Moreover, we have organized some methylated modification-related literature with a high IF for you. 1. Identification of Methylated Proteins in the Yeast Small Ribosomal Subunit: A Role for SPOUT Methyltransferases in Protein Arginine Methylation 2. Avenues for post-translational protein modification prevention and therapy 3. iPTMnet: an integrated resource for protein post-translational modification network discovery 4. SNF1-related protein kinase 1 represses Arabidopsis growth through post-translational modification of E2Fa in response to energy stress 5. Protein phosphatase 1 nuclear targeting subunit is a hypoxia inducible gene: its role in post-translational modification of p53 and MDM2 6. The functions and mechanisms of post-translational modification in protein regulators of RNA methylation: Current status and future perspectives 7. Actin R256 Mono-methylation Is a Conserved Post-translational Modification Involved in Transcription 8. Protein post-translational modification by lysine succinylation: Biochemistry, biological implications, and therapeutic opportunities.
|